I’m lookin’ at you, IDers 😉
Dembski’s paper: Specification: The Pattern That Specifies Complexity gives a clear definition of CSI.
The complexity of pattern (any pattern) is defined in terms of Shannon Complexity. This is pretty easy to calculate, as it is merely the probability of getting this particular pattern if you were to randomly draw each piece of the pattern from a jumbled bag of pieces, where the bag contains pieces in the same proportion as your pattern, and stick them together any old where. Let’s say all our patterns are 2×2 arrangements of black or white pixels. Clearly if the pattern consists of just four black or white pixels, two black and two white* , there are only 16 patterns we can make:
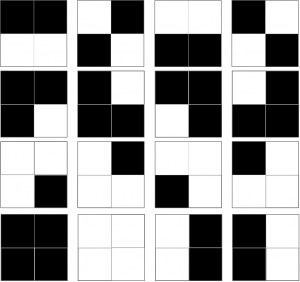 And we can calculate this by saying: for each pixel we have 2 choices, black or white, so the total number of possible patterns is 2*2*2*2, i.e 24 i.e. 16. That means that if we just made patterns at random we’d have a 1/16 chance of getting any one particular pattern, which in decimals is .0625, or 6.25%. We could also be fancy and express that as the negative log 2 of .625, which would be 4 bits. But it all means the same thing. The neat thing about logs is that you can add them, and get the answer you would have got if you’d multipled the unlogged numbers. And as the negative log of .5 is 1, each pixel, for which we have a 50% chance of being black or white, is worth “1 bit”, and four pixels will be worth 4 bits.
And we can calculate this by saying: for each pixel we have 2 choices, black or white, so the total number of possible patterns is 2*2*2*2, i.e 24 i.e. 16. That means that if we just made patterns at random we’d have a 1/16 chance of getting any one particular pattern, which in decimals is .0625, or 6.25%. We could also be fancy and express that as the negative log 2 of .625, which would be 4 bits. But it all means the same thing. The neat thing about logs is that you can add them, and get the answer you would have got if you’d multipled the unlogged numbers. And as the negative log of .5 is 1, each pixel, for which we have a 50% chance of being black or white, is worth “1 bit”, and four pixels will be worth 4 bits.
So if we go back to my nice greyscale image of Skeiðarárjökull Glacier, we can easily calculate its complexity in bits. There are 256 possible colours for each pixel, and there are 658 x 795 pixels in the image, i.e 523,110 pixels. If all the colours were equally represented, there would be 8 bits per pixel (because the negative log 2 of 1/256 is 8), and we’d have 8*658 *795 bits, which is about 4 million bits. In fact it’s a bit less than that, because not all colours are equally represented (light pixels are more common than dark) but it’s still very large. So we can say that my pattern is complex by Dembski’s definition. (The fact that it’s a photo of the glacier is irrelevant – the photo is just a simplified version of the original pattern, with a lot of the detail – the information – of the original removed. So if my picture has specified complexity, the glacier itself certainly will).
But the clever part of Dembski’s idea is the idea of Specification. He proposes that we define a specification by the descriptive complexity of the pattern – the more simply it can be described, the more specified it is. So “100 heads” for 100 coin tosses is far simpler to describe than “HHTTHHTHHTHTHHTHTHTHTHHTTHTTTTTH….” and all the other complicated patterns that you might toss. So if we tossed 100 heads, we’d be very surprised, and smell a rat, even though the pattern is actually no less likely any other single pattern that we could conceivably toss. And so Dembski says that if our “Target” or candidate pattern is part of a subset of possible patterns that can be described as, or more simply, than the Target pattern it is Specified, and if that subset is a very small proportion of the whole range of possible patterns, we can say it is Specified and Complex (because very low probability on a single draw, therefore lots of bits).
When I chose my glacier pattern I chose it because it is quite distinctive. It features waving bands of dark alternating with light. One way we can describe such a pattern is in terms of its “autocorrelation” – how well the value of one pixel predicts the value of the next. If the pattern consisted of perfectly graded horizontal bands of grey, shaded from dark at the top to light at the bottom, the autocorrelation would be near 1 (near, because it wouldn’t necessarily be exactly linear). And so we could write a very simple equation that described the colour of each pixel, with a short accompanying list of pixels that didn’t quite equal the value predicted by the equation.
However, for the vast majority of patterns drawn from that pool of pixel colours, you’d pretty well need to describe every pixel separately (which is why bitmaps take so much memory).
So one very easy way of figuring out how the “descriptive complexity” (or simplicity, rather) of my pattern was to compute the mean autocorrelation across all the columns and rows of my image. And it was very high. The correlation value was .89 which is a very high correlation – not surprisingly as the colours are very strongly clustered. And so I know that the simplest possible description of my pattern is going to be a lot simpler than for a randomly drawn pattern of pixels.
To calculate what proportion of randomly drawn patterns would have this high a mean autocorrelation, I ran a Monte Carlo simulation (with 10,000 iterations) so that I could estimate the mean autocorrelation of randomly drawn patterns, and the standard deviation of those correlations (for math nitpickers I first Fisher transformed the correlations, to normalise the distribution, as one should). That gave me my distribution of autocorrelations under the null of random draws. And I found, as I expected, that the mean was near zero, and the standard deviation very small. And having got that standard deviation, I was able to work out how many standard deviations my image correlation was from the mean of the random image.
The answer was over 4000. Now, we can convert that number of standard devions to a probability – the probability of getting that pattern from random draws. And it is so terribly tiny, that my computer just coughed up “inf”. However, that’s OK, because it could cope with telling me how many standard deviations it would have to be to be less than Dembski’s threshold of 1/1*150, which is only 26 standard deviations. So we know it is way way way way under the the Bound (or, if you want to express the Bound in bits, way way way way way over 500 bits).
So, by Dembski’s definition, my pattern is Complex (has lots of Shannon Bits). Moreoever, by Dembski’s definition, my pattern is Specified (it can be described much more simply than most random patterns). And when we compute that Specified Complexity in Bits, by figuring out what proportion of possible patterns the patterns that can be described as or more simply than my pattern, it is way way way way way way more than 500 bits.
So, can we infer Design, by Dembski’s definition?
No, because Dembski has a third hurdle that my pattern has to vault, one that for some reason the Uncommon Descenters keep forgetting (with some commendable exceptions, I’m looking at you, Dr Torley :))
What that vast 500 bit plus number represents (my 4000+ standard deviations) is the probability that I would produce that pattern, or one with as high a mean autocorrelation, if I kept on running my Monte Carlo simulation for the lifetime of the Universe. In other words, it’s the probability of my pattern, given only random independent draws from my pool of pixels: p(T|H) where H is random independent draws. But that’s not what Dembski says H is. (Kairosfocus, you should have spotted this, but you didn’t).
Dembski says that H is “the relevant chance hypothesis” (my bold), “taking into account Darwinian and other material mechanisms“. Well, for a card dealer, or a coin-tosser, the “relevant chance hypothesis” is indeed “random independent draws”. But we aren’t talking about that “chance” hypothesis here (for Dembski, “chance” appears to mean “non-design, i.e. he collapses Chance and Necessity from his earlier Explanatory Filter). So we need to compute not just p(T|H) where H is random independent draws: we need to compute p(T|H) where H is the relevant chance hypothesis.
Now, because Phinehas and Kairosfocus both googled my pattern and, amazingly, found it (google is awesome) they had a far better chance of computing p(T|H) reasonably, because they could form a “relevant chance [non-design] hypothesis”. Well, I say “compute” but “have a stab at” is probably a better term. Once they knew it was ash on a glacier, they could ask: what is the probability of a pattern like Lizzie’s, given what we know about the patterns volcanic ash forms on glaciers? Well, we know that it forms patterns just like mine, because they actually found it doing just that! Therefore p(T|H) is near 1, and its the number of Specified Complexity bits near zero, not Unfeasibly Vast.
But, Kairosfocus, before you go and enjoy your success, consider this:
- Firstly: if you had computed the CSI without knowing what the picture was of, you would have been stumped: without knowing what it was, there is no way of computing p(T|H) and if you had used random independent draws as a default, you would have got a false positive for Design.
- Secondly: You got the right answer but gave the wrong reason. You and Eric Anderson both thought that CSI (or the EF) had yielded a correct negative because my pattern wasn’t Specified. It is. Its specification is exactly as Dembski described it. The reason you concluded it was not designed was because you didn’t actually compute CSI at all. What you needed to do was to compute p(T|H). But you could only do that if you knew what the thing was in the first place, figured out how it had been made, and that it was the result of “material mechanisms”! In other words, CSI will only give you the answer if you have some way of computing the p(T|H) where H is the relevant chance hypothesis.
And, as Dr Torley rightly says, you can only do that if you know what the thing is and how it was, or might have been, produced by any non-design mechanisms. And not only that, you’ve got to be able to compute the probability of getting the pattern by those mechanisms, including the ones you haven’t thought of!
So: how do you compute CSI for a pattern where the relevant chance [non-design] hypothesis is not simply “independent random draws”?
I suggest it is impossible. I suggest moreover, that Dembski is, even within that paper, completely contradicting himself. In that same paper, where he says that we must take into account “Darwinian and other mechanisms” when computing p(T|H), he also says:
By contrast, to employ specified complexity to infer design is to take the view that objects, even if nothing is known about how they arose, can exhibit features that reliably signalthe action of an intelligent cause.
If we know nothing about how they arose (as was the case before KF and Phinehas googled it) you would have no basis on which to calculate p(T|H), unless you assumed random independent draws, which would, as we have seen, give you a massively false positive. You can get the right answer if you do know something about how it arose, but then CSI isn’t doing the job it is supposed to.
Conclusion:
CSI can’t be computed, unless you know the answer before you compute it.
* h/t to cubist. I changed my scenario mid-composition, but forgot to change this text!


And if this message-in-a-bottle bobs up against Eric Anderson’s boat, he might just like to pop the cork. He says:
“I knew somebody didn’t understand the difference,” said Rabbit, “and I knew it wasn’t me”.
Excellent post, Lizzie. I look forward to hearing the UDers’ responses.
Well, that seems very clear indeed, particularly as to what Dembski actually says; and I’d like to thank you, Liz, for taking the time to lay it all out like that.
I’ll be very interested in any response from beyond the pale
One small quibble. To calculate p(T|H), you don’t need to know how the pattern actually arose. You need to know all of the possible ways it could have arisen (by “Darwinian or other material mechanisms” , and the aggregate probability for all of them gives you p(T|H).
VjTorley’s argument is, in effect, that unknown mechanisms can be assumed to have a low probability, and can therefore be neglected. Your ash example shows why this is a huge mistake that leads to false positives for design.
Lizzie, is there a relationship between autocorrelation and compressability?
FOR RECORD: But of course, CSI , FSCIO, etc, are simply placeholders for “looks designed to me”, with a twist of arguments from large numbers without actual math. Shame on those who want the respectability of math without doing any.
I’m guessing that you could get lossy compressibility with auto-correlation but not lossless?
The way we thought CSI worked was that there was some scale, such as fitness or even Dembski’s algorithmic (non)complexity, and a pattern had CSI if it was so far out on that scale that to be that far or further was extremely improbable under a coin-toss (or pure mutation) model. Extremely improbable meaning having less than 10^-150 probability.
That was actually a reasonably meaningful definition — what was wrong with it was that there were no working conservation laws that said that natural selection could not accomplish that.
But that was before it was made clear to us all that we also had to compute P(T|H) for all relevant non-chance “chance hypotheses”. Now CSI is just a proxy for the conclusion of the Design Inference, rather than a method for making it.
Now the way that observing CSI rules out natural selection is that the user is asked to rule it out, and then only declare CSI to be present if natural selection can be ruled out.
On the issue of whether P(T|H) is needed, there are interesting disagreements over at UD between “joe” and Eric Anderson (on the one side) and vjtorley on the other. I’d also say, based on the quotations from him, between Dembski on the one side and Dembski on the other.
No foolish consistency, eh?
Seems most likely that Dembski noted that one needs information about the history of an object to determine its specification, because otherwise specification is impossible to assess. BUT then Dembski realized that if this is indeed the case, then life need not be Designed. In fact, in that case gods themselves are meaningless and superfluous!
So he asserts both sides. What else can he do?
Admit that his life work is rubbish?
Not really. See my comment above.
Ah, I see your point. Of course, one of the possible ways such a thing could happen IS its history. But I would categorize “knowing the intentions of the designer” as useful historical information.
Or looked at another way, “design” is being used as both a verb to describe a process, and as a noun, a result of that process. If you don’t know anything about the process at all (even the category of processes to which it belongs), then you can’t use the noun meaningfully.
How does one know all possible histories? Why would anyone take such a requirement seriously in a diagnostic test?
Beggin’ yer pardon, Lizzie, but you seem to have misspoke yourself here:
Given a 2×2 array of pixels, of which two are black and the other two white, there are only six “patterns we can make”. There are, of course, 16 different patterns one can make from four pixels, with each pixel being either black or white… but most of those patterns will not be evenly split between white and black. There’s the all-black and all-white patterns, plus the “3:1 split” patterns.
Judging by the diagram, I suspect it’s just a typo and that’s what she meant to say.
Yes, that’s a good point, and one I should have made myself. Thanks.
I feel like Dawkins reading quantum theory! It all makes sense when I read it …but five minutes later…. Looking at the responses at UD, there seems to be quite a bit of disarray. The point, made so elegantly, appears to have registered.
Gpuccio seems to make some kind of back-handed concession:
We can only deal with propositions at hand, gpuccio. If KF wants to write up something coherent about his own personal acronym – or if you want to try and quantify a property such as “functionality” – it will be reviewed with interest.
ETA link
Yes, I think so, but perhaps someone better at math than me could confirm.
I chose lag-1 autocorrelation as a crude proxy for low-frequency dominance – a pattern with a large positive lag-1 autocorrelation is going to be dominated by low frequencies. And low frequency vectors require fewer numbers to describe for a given time or space interval.
Or am I being naive?
You don’t, of course.
vjtorley tries to deal with the problem thus:
There are a couple of problems with that.
In the context of the ash example, this idea fails because one of “the remaining chance hypotheses” — ash deposition — turns out to be highly probable.
In the context of biology, the problem is that neither vjtorley nor anyone else can calculate the probability that a given instance of biological complexity is explained by the most probable “chance hypothesis” of all — modern evolutionary theory.
Show us the P(T|H), IDers!
Yes, it was a bad edit. I had originally meant to take the example of making patterns with just four tiles, two black, two white, then I changed it to a bag of tiles.
I’ll fix.
That’s interesting. On the contrary I’m perfectly happy with the concept of a functional specification, but clearly it’s inapplicable to this pattern (and would be for lots of patterns for which we might suspect design).
I think functional specification for proteins is a potentially viable concept – but all uses I have seen for it simply calculate something like FSC on the basis of p(T|H) where H is “random independent draws”. It’s a good enough way of describing the relevant space, and may, as even Durston et al say, be a useful measure when evaluating possible evolutionary paths that led to functional proteins. The Hazen et al (2007) paper, Functional information and the emergence of biocomplexity, is a good paper.
But no-one has shown that it’s “a good tool for detecting design”. All gpuccio has shown is that he has inferred design from finding proteins that exceed some value on such a measure. But as nobody is suggesting that proteins arose from random independent draws, if it’s calculated on the basis of H in p(T|H) being random independent draws, his inference is invalid.
As Dembski concedes, and as the false positive you’d get if you applied that H to my image demonstrates. You need to take into account all relevant non-design hypotheses, not just random independent draws, when calculating your null.
Which is why the whole idea is doomed of course. The reason we do null hypothesis testing in science is because it enables us to make predictions about our study hypothesis, H1, and compare our data with the probability of seeing the same data, given H0, the null. In general, IDist regard Design as the null – they consider that if we can’t explain X by material means, we must default to Design. However, that would mean making a positive prediction for Design, which they can’t do, because it would involve making some hypotheses about what a Designed universe would look like.
So Dembski had this neat wheeze of turning it round, and making “non-design” the null. But to make that work, he has to be able to calculate the probability of the data under that null. And to do that, as his “non-design” null consists of all possible hypotheses apart from design, he has to estimate what the probability distribution for the data would look like under all possible non-design hypotheses, including ones we haven’t thought of yet!
Which is clearly impossible. But it looked quite mathy, with bits and Kolmogorovs and all. As I said in an earlier post – it’s a great idea. Now bell the cat.
Tell us how to compute p(T|H), where H is “the relevant chance hypothesis” without smuggling in (heh) the answer you want as your assumption.
Methinks the man projecteth.
Golly, just read some of the more recent comments at UD.
They seem to be dropping CSI like a hot potato. They certainly don’t like Dembski’s definition of Specification, but of course they can’t say that (because my image meets it).
And I think that apart from Dr Torley, none of them get the point about p(T|H) needing to take into account mechanisms other than random independent draws, whether we are talking about a functional specification or any other specification.
KF simply took Durston’s FSC numbers, derived using H as random independent draws, and plugged them into his formula, without revising H at all.
They still can’t see the problem. Even though Dembski laid it out in black and white. It’s quite bizarre.
It’s as though the logic goes like this:
Oh boy, missed this:
Eric Anderson:
I guess Dembski’s Specification paper is fairly hard going, but from the guy who accused me of not understanding the difference between Specification and Complexity, it’s kinda funny. I know that Dembski’s mathematical definition of Specification is a bit of a crock, and Kolmogorov Compressibility isn’t a very straightforward quantity, but at least one can attempt a compressibility measure, as I did with my image.
Eric doesn’t even know that Dembski gave one! Well, Eric, it’s a shame you never click outside the UD bubble, but if you do accidentally find yourself reading this post, check out Dembski’s paper on Specification (the thing you think you understand and I don’t) and find his mathematical definition of Specification, which is low Kolmogorov Entropy, or high compressibility. My image is a low entropy image. It is therefore one of a small subset of the total number of possible images drawn from the same distribution of pixel values of my image with entropy as low, or lower, than mine.
If you think I’ve done the wrong math, you could show me how. But as you don’t seem even to know the math, I guess you can’t.
And in any case, you’d be better to concentrate on your p(T|H) term. That’s where the problem lies.
That’s good, but how much of the “non-ID” literature have you read, Eric?
I’ve sent links to the five relevant blog posts (UD and here) to Dembski, and invited him to weigh in, either here or at UD.
Dr. Dembski has not shown much enthusiasm in the past for engaging with critics, as I am sure Wesley Elsberry and Jeff Shallit would confirm. I wonder if Lizzie will be ignored. The current crop of commenters in the “CSI revisited” thread at UD could do with some support!
PS
I am reminded of the “explanatory filter” incident, where Dembski announced he was retiring the EF but, when his critics reacted with amusement, he reinstated it, to the extent that he has never mentioned the EF since. I bet, if Demsbki engages at all, that may be the scenario.
ETA
I was going to put a link but perhaps it’s off-topic so I’ll post it in sandbox.
It occurs to me that a possible rebuttal to my case might be this:
OK, so we don’t know what created this particular pattern, but the kinds of process that can create this kind of pattern includes a,b,c,d, and e, and many of these are non-design processes. And so even if this one was in fact designed, there is still a substantial probability that it came about by one of these non-design processes. Therefore p(T|H) where H is all non-design hypotheses is at least substantial. Do we do not infer design.
And I think that would be a valid use of CSI to reject a design inference in the case of my picture (even if it had been a butterfly wing – CSI is not claimed not to produce false negatives). That way you wouldn’t need to google the image first to retain the null of non-design. (Sometimes I think the DI should give me a fellowship.)
However: my counter-rebuttal is this: Let’s say we have a pattern that, like mine, is both Complex and highly Specified, but not one of a kind that we’ve ever seen created by any process that we know to be non-designed. And we have no non-design hypothesis. Do we infer design? Well, I don’t think so.
“We do not have a non-design hypothesis” is not the same as saying “there can be no non-design hypothesis”. For instance, for a long time, there was no persuasive non-design hypothesis for the pattern of “canals” on Mars. And many inferred Design. Eventually it turned out that there was a very simple hypothesis: the pattern was not on the surface of Mars at all – it was generated by the measuring instrument. And that hypothesis turned out to fit the data very well.. Not all measurement errors are “random”. A better example might be the axis of evil in the Cosmic Microwave Background. We have no non-design hypothesis to explain it. Can we infer Design? I don’t think so.
It comes back to the basic problem of null hypothesis testing. You have to be able to characterise the expected distribution of your data under your null.
If your null is “all non-design hypotheses” it must include hypotheses you haven’t thought of yet, you can’t characterise your null (at least Dr Torley addresses this, but for reasons others have given, I don’t think his idea works – there’s no reason that I can see to assume that hypotheses you haven’t thought of yet are less likely to give rise to your data than hypotheses you have).
But if your null is “Design” then you need to produce the expected distribution of data under that null, which IDists are loathe to propose, as it would mean making assumptions about what the putative Designer would be more, and less, likely to do.
Lizzie:
As you note, the real problem lies with the P(T|H) term, not the compressibility.
Dembski’s use of compressibility as the specification is inconsistent. In some of his works he declares that
(there is a similar statement in No Free Lunch).
In the same work quoted above he says that
So he is being clear there that he does not always consider compressibility to be the specification, just sometimes. And sometimes viability can be the specification.
But elsewhere he invokes compressibility as the specification. Which is bizarre if the objective is to explain how organisms can some to be of high fitness, since fitness is by no means the same thing as compressibility. Searching for genotypes that make high fitness phenotypes does not necessariy improve compressibility.
There can be organisms that are much more compressible than real ones, provided you don’t require them to successfully survive or reproduce.
I still can’t get beyond the simple question “how can you possibly say what function an unknown object might have?” Any calculation involving unknown quantities can hardly be used as a proof, can it?
Hope it isn’t off-topic but this seems almost prescient!
Jerry, a commenter at UD in 2008
From here
Jerry had part of the truth. ID hasn’t been fruitful full-stop.
Regarding the snark about the rest of the alphabet soup, I’ve googled for some definitions.
They all seem to suffer from exactly the same problem. The specification criteria are arguably less ambiguous (although how you tell a functional protein from a non-functional one that doesn’t actually exist, or out of the context of an actual organism, with actual tissues, in which that protein may or not be expressed, and in which environment such a protein may or may not be advantageous, I don’t know, but be that as it may….), but you are still stuck with the problem of how to compute p(T|H), if H is anything other than random independent draws.
And if H isn’t anything except random independent draws, it’s clearly a straw man.
Perhaps someone gpuccio or Kairosfocus could explain?
Or not. I think we can stick a fork in this one.
In the case of gpuccio’s “dFSCI”, he calculates the “functional information” of the sequence; that is, the probability that the sequence was produced by pure random variation with no selection.
To be considered dFSCI, the functional information has to exceed a threshold value, and it also has to satisfy a second criterion:
Thus, even if two sequences have identical, high values of functional information that exceed the threshold, they don’t necessarily both have dFSCI.
So P(T|H) factors obliquely into the assessment of dFSCI, but not into the numerical value associated with it.
And of course, the argument based on dFSCI is just as circular as the argument based on CSI. You have to know that a sequence could not have evolved before you attribute dFSCI to it, so stating that “X has dFSCI, so it couldn’t have evolved” is a mere tautology.
Yes. I think they are so focussed on the Specification problem that they don’t notice the eleP(T|H)ant in the room.
There are issues, but not insuperable issues, in defining Functional Complexity (fewer than for Dembski’s Specified Complexity, I suggest).
Characterising the null is a whole nother ballgame.
h/t my son
Love it! POTW!!
There are well developed theories for entropy in correlated random processes. I’ll just quote a few results for one-dimensional processes. Extension to random fields is non-trivial however the Russioan mathematician Dobrushin and others have developed useful results. For a one dimensional process say a discrete parameter process such as a time series the data can be written X(-1),X(0),X(1) …,X(n), …. If the X’s are independent and identically distributed the entropy rate (entropy per sample) is just the average of the negative logarithm of the probability densituy for X i.e. the usual formula.
For a Gaussian distirbution this is H= log(s) +1/2 log(2 pi e) where s^2 is the variance. When the process is correlated successive values can be partly predicted on the basis of previous values and for Gaussians processes the variance above is replaced by the prediction error variance which for highly correlated proecess is mush smaller than the raw series variance . Since the optimal linear predictor is recoverable from the autocorrelation sequence of the series, detailed numbers are easily generated for this case. An alternative recipe providing further insight is given by
log() => I nt{ log(S(f)) df }
S(f) = Sum(k=-inf,inf) R(k)exp(2pi *i*f)
R(k) =
where Int is the integral over frequencies (0,pi) for a real valued series S(f) is the spectrum which is just the fourier transform of the auto-covariance sequence and is the minimum attainable prediction error variance. thus as the spectrum is progressively confined to say, for iinstance, a set of narrow lines the predictiability increases and the entropy correspondingly decreases.
The bottom line is that any highly predictable process will have total phase volume that is minute compared to a purely random process of similar histogram so long as the data sample is large. If we take the correlation as the “specification” and the multivariate “null hypothesis” as independent identically distributed. variates of the same histogram (univariate probability) exhibited by the correlated process then Dembski would classify any such pattern as “designed”. He would argue that since the autocorrelation has to be retrieved from the data, that the specification is “cherry picked” and accordingly invalid, but to appeal to that he would have to abandon the use of algorithmic compressibility as being equivalent to specification since it is precisely by such (often implicit) retrievals that various compression algorthms proceed. Just knowing that the data is highly correlated could easily count as sufficient specification to exceed Dembskis bounds.
empaist,
Thanks! And welcome to TSZ!
The response at UD seems to be, however: focus on function.
Which seems sensible, given the problems with Specification. Unfortunately, they don’t stop with there.
Oops, spoke too soon. Looks like there’s a serious discussion of the math of Dembski’s Specification going on at UD.
I’d stick with function, guys. Seriously.
Function being entirely contextual, I’m not sure that would help.
They seem hung up on the enzymatic role of protein, and a ‘minimal function’ might be the ability to catalyse some reaction, somehow, in vitro. Stick it in an organism, however, and many products would fall at the first hurdle. You found the protein in an organism; your task is to determine how ‘unlikely’ it is. Given the precedent genome, how unlikely were the amendments that led to this one?
As to noncatalytic proteins that – for example – stabilise the ribosome, their function depends entirely on the presence and structure of the ribosome in question. One might be persuaded, by the present resistance to amendment of such proteins, that there is only one way to skin that particular kipper. But within your set of ‘chance hypotheses’, you have to consider co-evolution of RNA, protein and the cellular components which they make. Initially labile sequences can become pinned in place, because – in time – too much depends on them. Early ‘optionality’ can readily become modern constraint – such structural proteins are, indeed, much less capable of amendment than the average enzyme.
No, it won’t help. But at least they can concentrate on demonstrating that life is designed. Trying to decide whether glaciers or black monoliths are designed isn’t a major priority for ID, I don’t think, however Dembski defines it.
Interesting. Salvador Cordova is arguing that there are two versions of Dembski’s CSI, v1.0 and v2.0 (these correspond to the versions with or without the P(T|H) term). He is being reassured by vjtorley that there has never been any v1.0. There is also some further discussion at UD in the “CSI Revisited” thread.
The lineup seems to so far be:
1. CSI should be defined independently of mechanism:
Eric Anderson and Joe
2. CSI has always included the P(T|H) term so has never been defined independently of mechanism:
vjtorley
3. There are two versions of CSI, one like #1 and one like #2
Salvador Cordova
4. We should concentrate on versions of CSI that are defined in terms of “function”
gpuccio and kairosfocus
(who I think do end up including something like P(T|H) in their versions).
I think this list misses a number of other UD participants.
To recycle your excellent pun, is this a case of the blind men and the EleP(T|H)ant?
Oops, I posted the above comment in response to the wrong comment of Lizzie’s. It is in response to the remark about “serious discussion going on … at UD” which is below.
Indeed. Why is that not the starting point and not the end-point of ID “research”? The lack of curiosity baffles me. Let’s have some suggestions on how, when and where life was designed. If one assumes “life is designed”, what predictions can be made that would reinforce the hypothesis? Even the simplest questions such as what is the intervening interface between living organism [and designer]*, how often is intervention required, does evolution take care of the minor details or is it all design? Speculation and thought experiments are free. Surely some ID proponent can come up with a little more detail than “life is designed!”
*added in edit
Sadly, they can’t.
The Bible doesn’t provide any design details, why should they?
I have asked them at UD (never got any answers, though) how one determines function of a protein and then rules one is ‘more’ functional than another when considering the whole of protein possibilities. For example something so mundane as serum albumin where the principal function is its major contribution to maintaining osmolality where any protein in solution contributes to that same form of function.
BK,
Yes indeed. I saw that someone at UD had posted a video about Douglas Axe’s 2004 paper in the Journal of Molecular Biology (Estimating the Prevalence of Protein Sequences Adopting Functional Enzyme Folds). There was a good ,Panda’s Thumb piece on it, making much that point, as I understood it.