Dr. Winston Ewert put forward his module hypothesis, but I put forward an alternate module hypothesis at the domain and motif level of proteins. It is based actually on papers by evolutionists who have pointed out that the problem of “Promiscuous Domains” remains an unsolved problem in evolutionary biology.
When I put Promiscuous Domains on the table in the Common Design vs. Common Descent thread, the TSZ Darwinists ignored the problem and then declared victory. I viewed their non-response as evidence they didn’t understand the problem and/or preferred to ignore it.
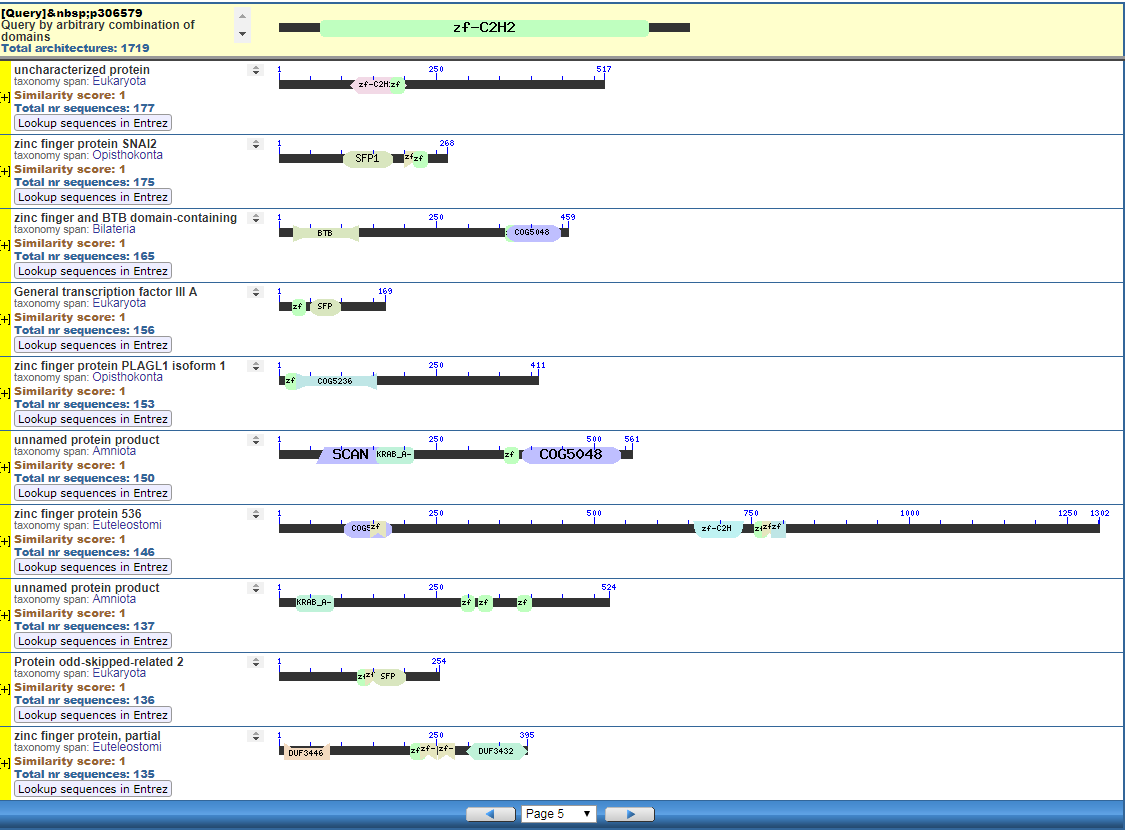
Perhaps pictures are worth ten thousand words. From the NIH, that great source inspiration for the Intelligent Design community, we have the CDART database viewer.
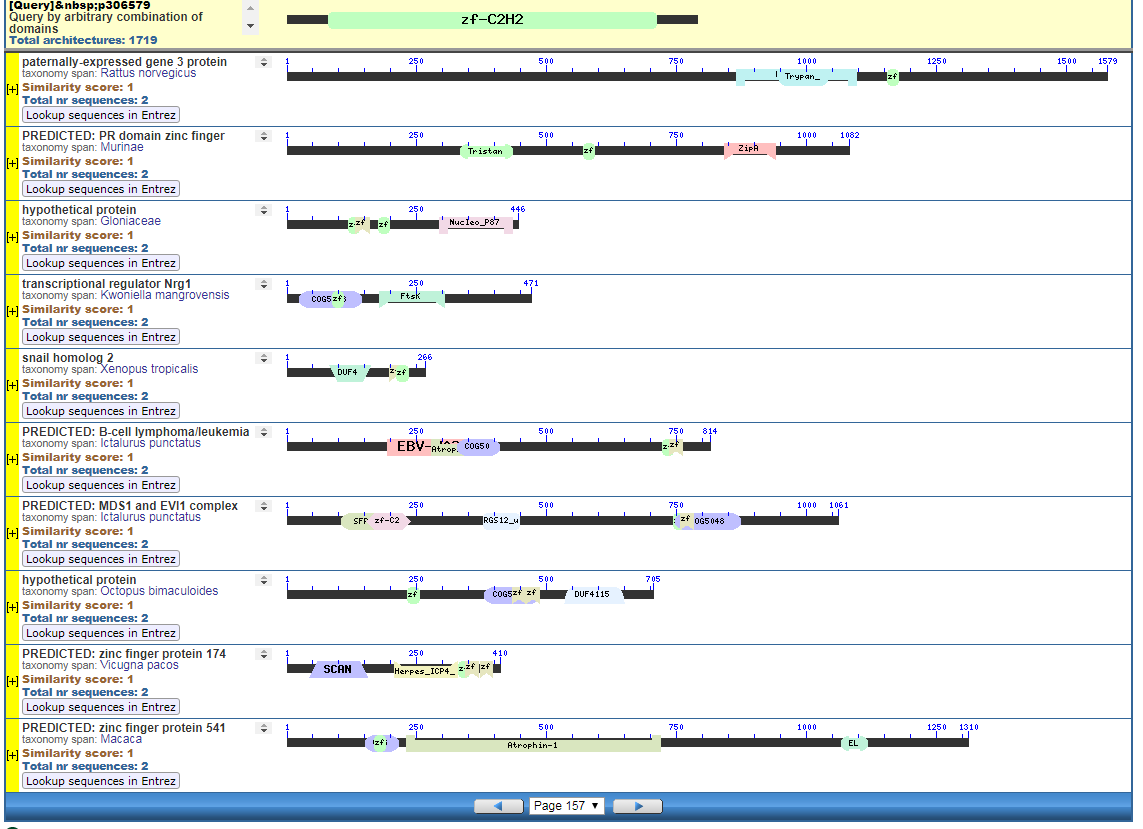
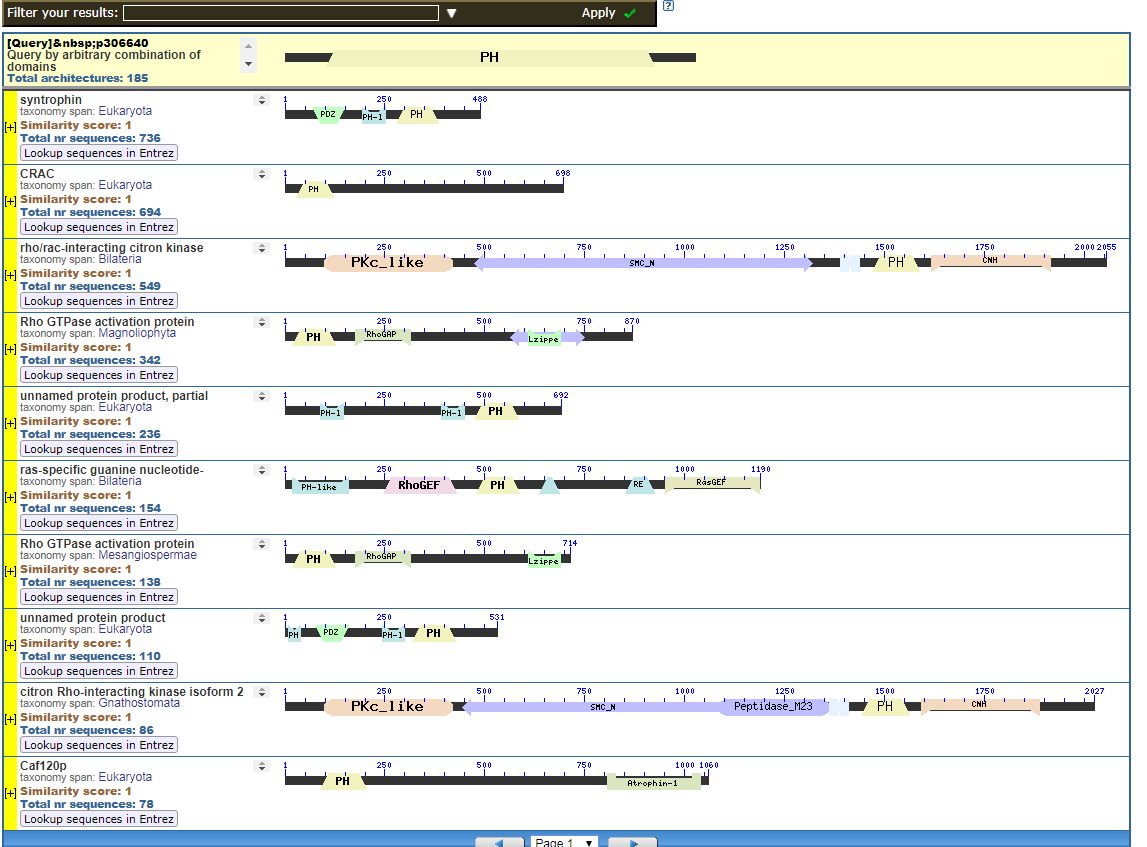
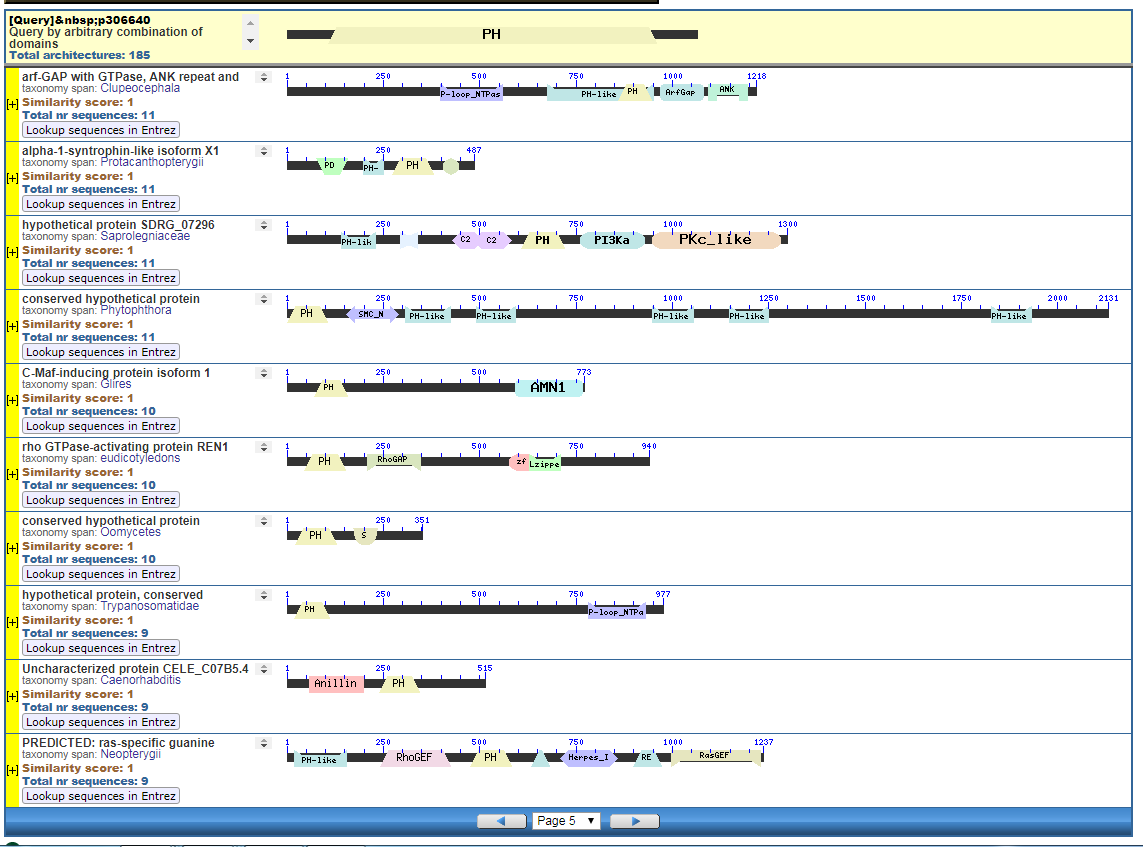
From the CDART viewer, I provide a few of the thousands of diagrams that show the promiscuity of protein domains. The diagrams below show the classical zinc finger ZF-C2H2 “ZF” domain and the Plextrin Homology “PH” domains. Note how the location of domains is “shuffled” to different locations in different proteins. It’s as if proteins are made by different lego-like parts in different order and position. My preliminary look into small 4-amino acid motifs that are the target of phosphorylating kinases suggests the the problem of promiscuity goes all the way down to small motif levels.
Such promiscuity is more consistent with common design than common descent.
Click to Enlarge Classical ZF-C2H2 Zinc Finger Page 5
Click to Enlarge Classical ZF-C2H2 Zinc Finger Page 157
Click to see all CDART Classical ZF-C2H2 Zinc Finger Architectures

How would you determine a heritable feature might have been miraculously created and inserted in a lineage so the hierarchy is preserved?
In principle, if you were there to observe the change with your own eyes and equipment over time, would that persuade you the hierarchical pattern evolved by common descent with miracles of special creation to tack on some heritable features?
Yes, no?
But since you weren’t there, hypothetically what patterns, if you saw them in extant organism might persuade you that even under the assumption of common descent, miracles of special creation were responsible for some of the heritable features?
Do you think a miracle was responsible for the creation of eukaryotes? Why or why not? If a eukaryote won’t persuade you of a miracle of special creation along the evolution of life, then describe what sort of creature would persuade you that evolved by a process of common descent with miracles of special creation along the way.
I’m going to hypothesis the KRAB containing proteins which appear only in tetrapods, which also have numerous promiscuous domains, are consistent with miracles of special creation. Whether they are special creations added to pre-existing organisms such that we have some sort of Universal Common Ancestry, or special creations of entire groups of species is a separate issue.
Statistically improbable “convergences” count as evidence for common design through acts of special creation. Convergence, is by definition, not the result of common ancestry.
But if statistically improbable convergences don’t count as evidence for common design, then what would?
I don’t know. It’s not something I’m all that interested in and, more importantly, it’s irrelevant to the subject we’re supposedly discussing here, which is common descent vs. separate creation of species. Because your language and the thought behind it are hopelessly confused, most of what follows is ambiguous and almost certainly off-topic.
Probably would, depending on just what you mean. But again, irrelevant to the subject.
Still irrelevant.
I don’t. I see no evidence of anything that couldn’t be explained by ordinary processes. But again, what does this have to do with the evidence for common descent vs. separate creation of species?
Exactly. It’s a separate issue, but it is in fact the very issue that your OP was supposedly about, right?
This is the most hopelessly confused pair of sentences in the whole rant. You are conflating “common design” of mutations with common design (=separate creation) of entire species. Evidence for the first, even if it existed, would not be evidence for the second. And still you continue to act as if it were.
I take it back. That was the most hopelessly confused sentence. You persist in throwing around ambiguous terms. If you hope to contrast common design with common descent you must define it as being in opposition to common descent, i.e. as separate creation of species (or of some groups or other), not as zapping of mutations into commonly descended lineages. I have concluded that the reason you keep changing the subject is that you are unaware that they’re different subjects.
Nested hierarchy shows common descent. Agree or disagree? That’s the subject.
I would say at any level, which is why scientists don’t claim such a thing. The “guidance” is provided by both negative and positive selection. Try and escape that false dichotomy. It’s not mere randomness versus god-did-it. There’s plenty of natural phenomena that have directions. Phenomena that are described in pretty much deterministic terms. I doubt you’d describe gravity as merely random, and I doubt you’d describe it as magical beings moving objects in accordance with their masses, distances, space-time-deformation effects, etc. either.
Nested hierarchy shows nested hierarchy. I demonstrated an intelligent agent can create sequences that follow a nested hierarchy.
But, since the topic is Promiscuous Domains, I’m going to discussion the possible convergence of domains. A convergence, is by definition not the result of common descent, right? So if a sequences converge on the same architecture, they converge on the same design, hence their similarity is by common design (be it through intelligence or some hypothetical mindless mechanism), not common descent.
The point of the OP was right there in the Title:
“Promiscuous Domains and Motifs Are Better Explained by Common Design than Common Descent, (Sal’s module Hypothesis)”
Soooo, we should be able to debate if something is a convergence. Whether it is because “God did it” or something else, is formally a separate issue, but a convergence is a common design. Even in the CDART database, they refer to domains as curated ARCHITECTURES!
Architectures are designs.
A zinc finger is a design, but it must be customized to the target. This is evident by attempt by humans to mimic the work of God:
https://www.ncbi.nlm.nih.gov/pubmed/16411739
Now that you know the terms of the OP, I hope we can explore specific protein sequences to argue for or against convergence.
How could we establish convergence as an explanation for the following patterns of zinc fingers?
Click to Enlarge
If the zinc fingers are mostly sequence identical, then one might argue the zinc fingers are due to exon shuffling or some duplication mechanism. But what if they are not sequence similar, how did the zinc fingers come to be in the first place on that protein. What did the common ancestor of these proteins look like since they only appear in tetrapods.
So when Tiktaalik rose from the sea, around that time, supposedly, these KRAB-containing proteins came to be. Right? How could we establish the sudden emergence of so many KRAB-conataing proteins? Would you assume only uber-KRAB-containing protein? Eh, that’s a problem because of KRAB is promiscuous!
You were the agent, if I recall, so some would doubt the demonstration. But more importantly, nobody doubted that an intelligent agent could create a nested hierarchy. An intelligent agent could create any pattern at all, and thus what one could do has no value in a scientific hypothesis. One would have to argue about what an intelligent agent would do, as you have done when it suits you. So why would an intelligent agent create a nested hierarchy, specifically the one we observe in life?
Here you play with ambiguity in the word “design”. Are you trying to ensure that nobody ever knows what you mean by any word?
I will speculate that nobody who read that title had any idea that by “common design” you meant nothing more than “convergence”. Including you, until just now. You seem bent on removing any meaning from your words.
So, are you abandoning your attempt to show that nested hierarchy is not evidence of common descent? Will you agree that convergence is not evidence against common descent of species?
stcordova,
Sal, have you ever looked at the scientific literature to see if any of your questions have been answered? A quick google search on “evolution of KRAB-containing proteins” produced a number of papers on the subject. Have you ever looked at any of them?
Entropy,
How do you account for the origin of gravity? Did it emerge by accident?
colewd,
I will ask it differently. How does inheritance (common descent) explain why these species have differences?
I don’t know if it even emerged Bill. If I had to guess I’d think of it as part of the nature of space-time-mass-energy. Part of the fabric of the cosmos, as some would say. In other words, I’d think of it being part and parcel of the universe, not something that would “emerge” independently of it. But I don’t really know. I’m talking from the little knowledge I have about gravity.
But aren’t you missing the point? Did you understand the problem with that false dichotomy?
Entropy,
I understand the potential for a false dichotomy. No one has shown that environmental forces can have a measured effect on the direction of mutations toward improved fitness except for some serendipitous short term gain. I think at best it slows down genome degradation.
I agree with you that nature by itself is not random.
What points to creation in my opinion is the origin of a highly predictable entity leading to species that can observe it.
The differences are explained by the nature-of/tendency-towards divergence. Neither by inheritance, nor by common ancestry. Divergence.
It doesn’t. It explains the pattern of distribution of the differences. Why they have differences is presumably related to how and why mutations and fixations happen, which, once more, is not explained by common descent.
John Harshman,
I agree that mutations and fixations explain some of the differences.
I don’t agree you can explain the hierarchal pattern with common descent alone. You have to explain the differences.
I agree that common descent is a plausible explanation for the similarities.
I think you can observe the pattern of distribution of the differences but to explain the pattern you need a mechanism.
Common descent can explain the pattern of similarities as we know the mechanism of inheritance passes genes from one generation to the next.
Since John Harshman and company are so certain of universal common ancestry based on hierarchical patterns, the KRAB proteins should be nested in something else, right? Yes? No?
How would one determine where KRAB-containing proteins nest such as the one in the 2nd row below, the ZNF136 protein?
Did that ancestor of ZNF136 have 14 zinc fingers in it to start at the origin of life? The ZNF136 protein has at least two domains, KRAB and 14 zinc fingers. So what did the common ancestor of them look like. Were the zinc fingers emerging by common descent or convergence.
This is hopelessly confused, as is so often the case with you. The pattern of similarities and the pattern of similarities and differences are the same thing. At a given site, some species could have C, while the remaining species have T. C is a similarity among those species that have C and a difference between them and those species that have T. Every similarity is a difference too. And, in a nested hierarchy, the distributions of both C and T are explained by the same thing: either at some point a C changed to T or T changed to C. Common descent doesn’t explain the change. It explains the distribution of C and T among species. Common descent with branching is the mechanism that explains the distribution of similarities and differences among species.
As I predicted, you ignored almost everything I said.
Can’t answer because I don’t know what you mean by “the KRAB proteins should be nested in something else”. Are you perhaps confusing common descent of species with common descent of different genes within a genome?
What do you mean by “where proteins nest”?
Ah, so you’re talking about common descent of genes within a genome. I think. You can do phylogenetic reconstruction by looking both at the different genes and at the genomes of many species. Thus you can trace paralogous genes to their point of duplication and reconstruct ancestral states of genes and proteins. Of course this is easiest with whole-gene duplications. If there’s exon- or domain- or motif-switching it becomes harder. There is no particular expectation that any of these genes will stretch back to the common ancestor of all life. But that’s also something you can investigate using phylogenetics.
Have you looked at any of the literature on the evolution of these proteins, as I suggested? It would seem the place to start if you’re looking for an answer to your questions. Of course we both know you aren’t actually looking for an answer; you’re looking for anything that fits the answer you have already decided on and will ignore anything that doesn’t fit.
Sal, try this: http://dev.biologists.org/content/144/15/2719
For one thing, KRAB proteins don’t occur only in tetrapods. They occur in all sarcopterygians. They are all probably homologous, with lots of gene duplication and loss. And it appears that the ancestral protein may have had a single exon. See? You should check the literature.
Thanks a million!
My point is that an expert biologist in a graduate program such as yourself shouldn’t have to be spoon-fed the literature. You should have thought of it yourself. Why didn’t you?
John Harshman,
No, it does not explain the distribution because different genes are distributed and does not explain why the different genes are part of the distribution.
It only explains why the same genes are part of the distribution. You are using rhetoric instead of clear scientific thinking. You cannot explain the nested structure with common descent on its own.
From my seat it looks more like you’re not understanding John’s example, and thus you conclude it’s rhetoric. I’d advice to focus and see where you might be a tad confused, and ask for clarifications (I understood because I know exactly what John was talking about, so I picture a multiple alignment and the Ts and Cs in a column, etc, which is something you most probably have no experience about).
Similar confusion happened to me with that comment where you were saying that my points were good, but I didn’t get it and thought you were complaining is some obscure way. I had to re-read it to realize where my confusion was.
Well, you have to be willing to try and understand, and to admit that you might have misunderstood some explanation.
Because he’s stuck in creationist-bullshitter mode. All he knows about literature searching is that it helps the quote-mining procedure.
Sigh. More word salad. “Different genes are distributed”? “Why the different genes are part of the distribution”?? It’s impossible to respond when neither of us has any clue what you’re talking about.
John Harshman,
Only where the genes are the same among T species. It does not explain C is in some species and not in others. You need to invoke something else to explain this.
Entropy,
If you can clearly show how common descent on its own explains differences in the tree please go for it.
I told you already that it doesn’t:
And so did John:
Because I have you to help me. I’m a molecular biophysics guy, not a phylogeneticist.
And surely “biophysics guys” don’t need to know about searching the literature! Really John, what’s wrong with you?
Entropy,
Divergence is different then common descent so you are agreeing with me. We need to invoke the mechanisms of divergence to explain the hierarchal structure.
I don’t know if I’m agreeing with you. As I said, John told you the very same thing that I told you, with different wording.
Either way, we need to put the whole thing together to explain the hierarchical structure.
1. Divergence happens somewhat constantly. Generally-speaking, the more time, the more divergence.
2. If populations separate their paths, then they will diverge from each other after such separation.
3. Thus, populations separated at different times will show different amounts of divergence.
4. If there’s common ancestry between species, their amounts of divergence should relate to the time since separation. Recent separation would mean less time, less divergence.
5. If there’s common ancestry between species, then the patterns of divergence should form hierarchies, with a subset of similarities within a closer group being somewhat “constant” but different to those within a separate group (the similarities should tend to be “nested”). So, for example, in some position, group 1 would tend to have a T, while group 2 would tend to have a G, indicating that the T belongs to group 1, and the G to group 2. Farther organisms would show variability in that position for this example. It’s a bit more complicated, and we’d have to consider many more positions in a multiple alignment, but the Ts and Gs would be thus “nested,” with the explanation being that the common ancestor for group 1 had a T, and the one for group 2 had a G.
So, it’s a combination of the tendency towards divergence and the times since the populations forming different lineages separated. Common ancestry and divergence, plus the patterns of divergence. Not common ancestry alone.
I hope that came clear enough, and that John will not feel offended by my oversimplifications and/or oversights, which I hope are not too damning.
Sigh again. Word salad again. You appear not even to know what T and C mean, which was obvious to anyone reading who has any clue about the subject at hand. However, though I can tell that you don’t know what you’re talking about, I can’t tell what you meant to say, and so still can’t respond.
C and T, incidentally, are two nucleotides at the same position in homologous DNA sequences in some set of species. In other words, we’re talking about a single point mutation. Do you know what nucleotides are? Do you know what point mutations are? There are no “T species”, and “the genes are the same” makes no sense. And yes, common descent (on some particular tree) does explain why C is in some species but not in others, because the species with C inherited it, and the species with T did not.
Entropy,
It is clear and I agree with you that ancestry (common descent) and divergence are components together that can build a nested hierarchy at the sequence, gene and morphology level. The debate is how much common descent is involved and what are the causes of the divergence.
Hey, common ground 🙂
What do you mean by that? Can you be more specific?
John Harshman,
At one extreme universal common descent and the other extreme, every specie was specially created.
The other possibility is the kind concept where there are created kinds but speciation among kinds. Crocs/Alligators could be a kind, or dogs/wolves could be a kind.
If there were multiple separate kinds, wouldn’t there also be multiple separate nested hierarchies? That is, nested hierarchy within kinds but no such structure between kinds? If you disagree, you’re back to having to explain why there should be nested hierarchy between kinds. But if you agree, that should be a way to discern kinds.
John Harshman,
As I have stated before I think with components from genes to tissues to complex structures like circulation there is a reuse of components and I don’t see any reason that a normal design process (as we know from software or computers) would not create a hierarchal structure. I would expect that created kinds if we can identify them would have very little hierarchal structure except at the sequence level.
I think this is an interesting point of discussion going forward.
Isn’t that just one of those assertions you are fond of accusing me of making? Can you support that idea beyond “I don’t see any reason”?
Again, it isn’t at all clear what you mean. Hierchical structure within kinds or between kinds? Why is the sequence level an exception to this lack of structure?
“I don’t see why…” is not an argument. Your inability to figure out why does not somehow constitute a valid reason to think that it would. You have to do the work and show with concrete examples that nesting hiearchical structure follows from re-use of parts.
And particularly it would be interesting if you could show that different parts getting re-used in different species yield the same hiearchy. (That’s the consilience of independent phylogenies bit).
We have been over this back here, where it was obvious that your intuitions failed you at every level. By now it is pretty much established that what you think should or shouldn’t follow is not a reliable guide to truth as your ideas just keep coming up wrong when put to a test.
I’d like to thank all who have participated in the discussion so far.
For the time being, I’m simply going to reposit here in this thread data I’m gathering along two lines:
1. promiscuous MOTIFS associated with human TopoIsomerases that are likely targets of post-translational modifications PTM, particularly phosphorylations since there is a lot of data on phosphorylations compared to other PTMs.
2. Zinc finger proteins, particularly those containing the KRAB domains
Thanks again in advance to all who are participating.
The Uniprot Topoisomerase 2-alpha data is here:
https://www.uniprot.org/uniprot/P11388
Starting at position 1501 is this sequence:
DFDSAVAPRA
The “S” is listed as a phosphorylation target. I strongly suspect that Uniprot may not capture all phosphorylation targets, but that is one that is at least listed.
The question is what sequence in that string is the MOTIF that is the target of the Kinase? Hypothetically it could be all sorts of things like (using UPPER CASE to emphasize the potential motif):
DFDSAVAPRA
dfdSAVApra
DFDSavapra
dFDSAvapra
dfDSAVapra
etc.
And that only assumes the target only 4 amino acids long.
I’m going to guess Sava. However phosphosite.com lists both dfdS and Sava as valid potential targets, but does not identify the kinase. This phosphorylation does not appear in rat or mouse Top2As because there is no Serine residue at that position. The serine appears in Dogs however.
There are possible methods to estimate the actual kinase target and thus we might actually be able to predict what the kinase will look like based on the target, but this could also be inferred from the hierarchical pattern which I suggested is not due to common descent, but rather an optimal method the Designer put in place to facilitate scientific discovery.
I’ll post some of my findings on this here on this thread.
Public masturbation is in poor taste.
John Harshman,
I would expect sequences to diverge by random mutation and fixation between species in the same kind. I would not expect new anatomical structure or new genes except for possible rare events.
At this point I want to hold off defending the claim that design can create the similarities we see as this is a differences discussion that Salvador is having.
As usual, you answer one bit and ignore the rest.
Whatever do you mean by “a differences discussion”? And Sal isn’t having a discussion at all. He’s using this thread, apparently, as a storage place for random crap he finds on the internet.
stcordova,
Why discovery vs simply a change in function?
.. completely without any evidence or even ability to test. But you suggested it, and that’s good enough for creationists who will proceed to believe mere suggestions, in the complete absense of evidence or even a method of testing it, with a psychotic level of conviction.
Well, actually the idea was given a blessing by a PhD dissertation committee of some unnamed individual (not me). I’m merely building upon a great idea. 🙂
I suspect in due time the unnamed individual will be made known.
Besides, at this stage in this thread, I’m just reporting what I find, I’m not doing advocacy work. Advocacy is reserved for in person speaking engagements for the most part, like Joel Tay (linked earlier in this thread).
Below is the protein interaction map of the Alpha Isoform of Topoisomarse in Humans. I post it to contrast it to the Beta Isoform in the next comment. Sequence alignments are strongly suggestive of why the Alpha and Beta Isoforms interact with the rest of the body differently. This interaction map is for skeletal muscle cells.
Note the 4 kinases: PKCg (protein kinase C gamma type), PKCa (protein kinase C alpha type), ERK2 (Mitogen-activated protein kinase 1), CK2a1 (Casein kinase II, alpha chain), PKCb (protein kinase C beta type).
This is NOT the same set of kinases that interact with Topoisomerase 2B (beta). Not to mention, the other sets of proteins that interact differently between Top2A and Top2B.
The point? It would seem the difference in the isoforms really matter! Random mutation creating this level of specificity? Nah….If there is common descent, it was intelligently designed.
Click to ENLARGE
The next is an interaction map of the Topoisomerase 2B (beta) isoform. Note that unlike it’s alpha counter part it has a different set of kinases targeting it, in fact only one, the ERK1 Kinase (Mitogen-activated protein kinase 3), which is different from the ERK2 Kinase that targets Top2a.
Did random mutation create such specificity of interactions (and hence specific amino acid targets/logos)? Doubtful.
Click to ENLARGE
Sal in a nutshell: Post a pretty graph or picture, make a generalized statement with some appreviations sprinkled over it, then declare something completely unsubstantied that doesn’t follow from any of the stated facts.
That’s right.
That’s wrong. The point of the graph is to point out the sequence differences, right down to even 4-amino acid motifs, HECK, even a single amino acid (like a Serine or Threonine) in the right location makes a difference. That’s not consistent with random mutation in the process of common descent, at best it is consistent with little acts of special creation with common descent, but if one starts invoking little acts of special creation, then what’s the use of common descent, the hierarchy is just as consistent with special creation by a Designer who delighted to build things with hierarchical relationships to facilitate scientific discovery.
I demonstrated this with the critical metal binding proteins and two phosphorylation sites (one in Top2A and one in Top2B). There will be more to come.
Bottom line: either you think the patterns of similarity and diversity are the products of mutational accidents, or they are the products of a design. I’ve provided evidence that even small motifs cannot be accidental because it would compromise function.