One of the most brilliant evolutionary biologists of the present day, Richard Sternberg, PhD PhD was ousted and permanently blacklisted by the National Institutes of Health and the Smithsonian Museum for his ID sympathies.
Sternberg is neither a Creationist nor Darwinist but classifies himself as a Process Structuralist which means he is not much involved in the ultimate questions of how things came to be, he just appreciates the amazing patterns of similarity and diversity in biology.
He was labelled by some of his former supporters as an intellectual terrorist after he used his position as editor of a journal to publish an ID-friendly article by Stephen Meyer in 2004. He paid dearly for that decision, and his subsequent dismissal from the NIH and Smithsonian precipitated special investigations by members of Congress and the White House a decade ago. Unfortunately, nothing of consequence was done for Sternberg and he was destroyed professionally and personally.
Despite his circumstances, he continued to publish excellent essays such as the one that highlights the non-random patterns of SINES (presumed by some to be junkDNA) which are present in mice and rats (link below).
To understand his essay, I will describe the essentials of his essay with a parable. Suppose we had two mostly identical stories published. The stories are identical except for the fact that in one version of the story the name of the main character is “Mary” and in the other version, the name of the main character is “Caroline”. Even if the two versions of the stories were generated from the same template, the changes in the two descendant copies could not have been random.
So even if we assume some sort of common descent of the two versions of the story from some ancestral copy, the differences in the versions could not be the result of random copying errors, but very deliberate and methodical changes in the duplication process that created the two versions. This peculiar phenomenon plays out approximately in the genomes of mice and rats, and I call it the Sternberg-Collins paradox in honor of Sternberg who brought the paradox into prominence and Francis Collins who was among the first to comment on the anomaly.
Assume for the sake of argument rats and mice came from a common ancestor. Are there differences which are non-random and thus evidence for non-random mutation? Sternberg effectively answers, “yes”.
The mouse and rat genomes look very similar, but there are sequences that repeat over and over again in each of their respective genomes and mostly in the same corresponding locations. If these sequences were identical in both lineages, there would not be much of an issue, but they are different even though they are in the same general corresponding locations.
Let me call one set of these repeating sequences in the mouse genome “Mary” and the corresponding sequence in the rat genome “Caroline”. The name “Mary” appears in numerous places in the mouse genome, and in the corresponding places where “Mary” appears in the mouse genome, “Caroline” appears in the rat genome.
Of course this was a figurative way of describing what is going on. “Mary” is in reality the B1/B2/B4 set of SINE retro elements in mice, and “Caroline” is in reality the ID SINE retro elements in the mouse. But don’t get hung up on the fancy language, the basic problem of non-random changes from a supposed common ancestor is brutally evident. A graph that shows the non-random changes is here and explained in Sternberg’s essay:
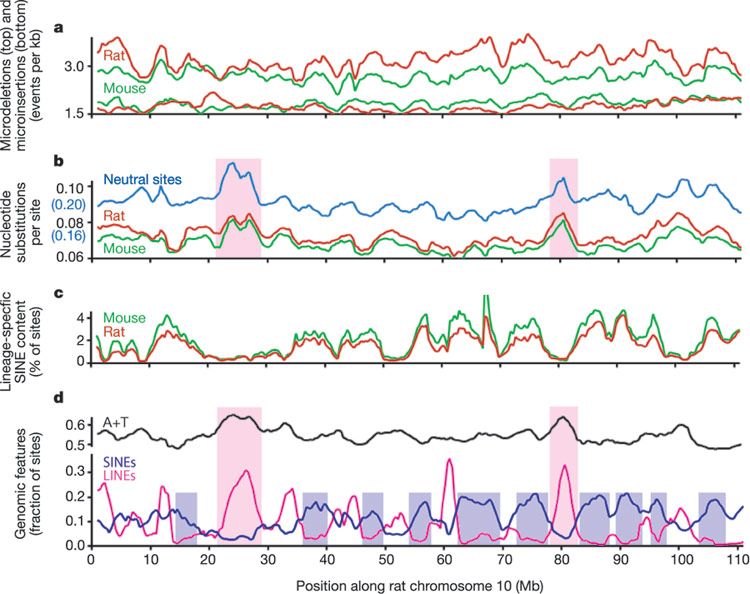
Sternberg’s point is that if mutational events happened, it was non-random, since to suppose it was random is an absurdity in the extreme. It cannot be the result of random DNA copying errors, but some non-random copying mechanism if common descent were true.
Is the non-random pattern the result of natural selection? Sternberg doesn’t address that question. But if the non-random SINE pattern is the result of selection, then this would mean the SINES aren’t junkDNA.
But if natural selection is assumed as the mechanism, there would be issues of the evolvabilty of so many nucleotides simultaneously. We’re talking maybe 300,000 SINE insertions in each lineage! For mice, the B1 sine is about 150bp and homologous to the 300bp primate Alu. The B2 SINE is 190bp. I could not find the size of the rat ID SINE nor the mouse B4 SINE, but I presume they are within the range of most other SINES (75-400 bp).
The source article in Nature which Sternberg referenced and had a buzzillion co-authors can be found here:
Genome sequence of the Brown Norway rat yields insights into mammalian evolution
That article points out:
Despite the different fates of SINE families, the number of SINEs inserted after speciation in each lineage is remarkably similar: approx300,000 copies.
Sternberg highlights the issue of having 300,000 non-random insertions happening in parallel in the two lineages after they split. He lays out the paradox which is the focus of this OP here:
Beginning to Decipher the SINE Signal
FWIW, I suspect there are probably similar issues with the Alu elements that appear in primates. Certainly this would be an issue if the mouse B1 (homologous to primate Alu) is in homologous locations in the primate genome. If that is the case, the Sternberg-Collins paradox is in play as well for primates. Perhaps one day we’ll know for sure if this is the case.

Yeah, I am very familiar with that work. Turns out this is classic God of the Gaps reasoning. Since he first pointed this out, we have figured out the mechanisms that give rise to this pattern. It just takes a bit of biological knowledge and PubMed to understand it.
Dr. Swamidass,
We’re very very honored to have someone of your caliber join the discussion. Thank you for visiting.
respectfully,
Sal
Wow, what a pleasant surprise. I’m used to being flamed on the internet by ID people. Thank you for being kind. I’m genuinely suprised.
swamidass,
Well, I think I heard your name mentioned when I was having beer (actually coffee and pastries) with Paul Nelson and Kirk Durston. My understanding is you are a Christian and also a researcher of well-respected ability.
I used to be a theistic evolutionist, I changed my mind somewhere along the way.
As far as Sternberg’s assertion, there is no mention really of God, he was pointing out the non-random nature of the insertions.
I posted this not to argue God of the Gaps specifically, but to put the issue of non-random insertion mutations as being real. Sternberg himself suggested isochore theory.
I’m not so sure one can say we have uncontested mechanistic explanations that have been confirmed in a laboratory context that SINES will retrotranspose with sufficient preference to create the non-random patterns. I suppose if that were the case we wouldn’t be so concerned with the medical impact of bad Alu insertions creating birth defects.
I’ve read papers on the matter, and I don’t see what you’ve referred to, but if you have specific citations, that would be helpful for the readers and myself. Thank you.
?
Readers may be interested in the somewhat more balanced view of what actually happed published by Wikipedia
https://en.wikipedia.org/wiki/Sternberg_peer_review_controversy
Why are we assuming this “for the sake of argument”? Is it your contention that rats and mice didn’t (or may not have) come from a common ancestor? If so, what’s your alternative and what evidence do you have for it? If not, why the qualification?
Well that is very kind of them. In private, I have tussled with both of them about the science, and especially in the case of Kirk, I have not always been charitable. So kudos to them for being generous to me.
swamidass,
swamidass,
Aren’t rats and mice of the same “kind” according to creationists?
Stopped reading right there. The rest has already been dealt with before.
If that was all that Sternberg did, I would have no dispute. In fact, he has done much more than this. I took exception to the original essay’s (accessible here,
http://www.evolutionnews.org/2010/03/signs_in_the_genome_part_2032961.html) insinuation that…
1.Theistic evolutionists are not being honest about the data. He writes, “what I am saying is that we know a lot about the genome that is being glossed over in the popular works that the theistic evolutionists write,” which is a very strange accusation. Popular works always gloss over details and no one (not even Collins) denies that there are interesting patterns in genetic data.
2. Somehow we need to look “Outside the BioLogos Box” (to ID?) to understand this pattern in the data. This is why I call it a God of the Gaps argument. Perhaps you are not making it, but it seems like Sternberg is. He makes this clear in the follow up article http://www.evolutionnews.org/2010/03/beginning_to_decipher_the_sine032981.html, that isochores could only be part of the answer, and he implies that this is evidence of God’s tinkering; “Isochores might provide a clue to cause of the mystery signal, but the cause–whatever it may be–is outside the BioLogos box.” In the context of this whole discussion, he is making an obtuse reference to ID.
Putting aside the implications of dishonesty he makes, this type of reasoning is never terribly strong in science. There are a few big scientific errors he makes:
1. He seems to be unaware of how SINES and LINES are inserted. This is actually is what leads to the correct answer (not ID, but a specific biochemical mechanism). Ignorance is fine, I do not fault him for it, but it makes for a horrible scientific argument.
2. He has what appears to be a very different understanding of “random” and “pattern” and “non-random” than is commonly used in science. I’ve noticed that ID thinkers often equate random with “uniformly random”, where biologists use this in a more precise sense. We think randomness can have distributions that deviate from uniformity (e.g. we call a gaussian distribution random). Identifying pattern in the data (as is done in the Collins papers) it is just to say that there is a clear deviation from uniformity, which indicates there is some mechanism underlying this process that deviates from uniform randomness. So, when we report “patterns” in the data, this is immediately translated as “non-random” in some ID people’s heads, when all that should be taken is that “there is a pattern in the randomness that deviates from uniformity.”
3. Any how, it turns out that there is a very good reason for this deviation from uniformity, a specific biochemical mechanism. It turns out, we do not need to go outside the “BioLogos Box”. In fact, attempting to look for ID here, just clouds our thinking and prevents us from seeing the obvious answers.
Now, I will point out, as a trained Biologist, I knew immediately what the explanation was likely to be. It had to be something tied to the TE insertion mechanism, which involves a class of enzymes (transposases, https://en.wikipedia.org/wiki/Transposase, https://www.ebi.ac.uk/interpro/potm/2006_12/Page2.htm) that catalyze SINE insertion, and is often relatively independent of SINE sequence. SINE insertion distribution is more correlated with the biochemical behavior of the transposases, which is not easily understood from the transposase sequence, is not necessarily dependent on SINE sequence, but often is connected DNA structure.
Apparently not aware of this mechanism, Sternberg jumped to the false conclusion that SINE insertion distribution was defined by the SINE sequence. So they conclude, therefore, there is no explanation for this in the BioLogos box. For a biologist, this really a bizarre logical jump that makes no sense in light of what we know of transposons, how we think of DNA, and how scientific inquiry progresses.
So knowing these facts, the most likely cause is a similarity in the rat and mouse genome sequences in concert with transposons with similar behaving transposases (not similar SINE sequences). The most likely possibility in the genome sequences, with that pattern, was probably DNA structure. With some focused googling, I quickly found this paper:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC383295/
(Shan Yang et al., “Patterns of Insertions and Their Covariation With Substitutions in the Rat, Mouse, and Human Genomes,” Genome Res. 2004 Apr; 14(4): 517–527.)
I should add that this is not argument against the function of every single SINE element. Clearly, there is strong evidence that some are functional (the most obvious case is where they insert into an exon). Alu’s also have been shown to carry regulatory elements (which we are certain has driven evolution at times). However, it seems like most TEs are not functional. Regardless, Sternberg’s reasoning here, to me, seem deeply flawed.
I should also add that some of the best evidence for Common Descent come from SINE and LINE elements. And the data Sternberg refrences is no different.
http://www.thegospelandevolution.com/StoryInOurGenes/audio-slides/2of2/
Anyone else think it’s funny that Sal thinks random means “equiprobable distribution” ?
It’s a very common ID / Creationist misconception – a silly position for a seasoned blackjack player (Hi from Reno).
Rumraket,
What I think is funny, is that you think anyone who doesn’t share your worldview is not capable. Laughable actually. Preposterous even.
God must have been so so mean to you.
I’m not claiming Cordova is making this error. I don’t know. Sternberg, however, clearly is.
swamidass:
Cordova (aka ‘Sal’) is making the same mistake:
Maybe you care right but let’s see if he agrees and what he discloses.
To clarify a possible misconception here, we aren’t talking about parallel insertions at specific sites in mouse and rat, but about parallel densities of insertions in large genomic regions. Sal, do you agree?
That’s not true, I don’t believe that, never have. You’re welcome to try to cite where I ever said that. You won’t find anything because you’re attributing to me an argument I’ve never made.
Probably, but so what?
Even if they share a common ancestor (and I accept that they do) the change in the two lineages is non-random. Darwinism continues it’s agonizingly slow death, but like other 19th century science it too is being replaced. Time for the skeptics to get on board!
Context matters, Rumrickety.
What ever Sal thinks about this is really beside the point any ways. I’m willing to take him at his word.
Regarding the science here, all we see is a pattern in the data.
One reason for that pattern is functionality, as Sal has suggested. Another reason for this pattern is common biochemical mechanisms (which never work by uniform distributions on DNA) and common evolutionary history (which leaves a very strong signature in genomes). Because multiple processes leave the same signature in the DNA (at the resolution we are discussing), the existence of a pattern alone really tells us nothing about function. Absolutely nothing.
To get to function, we would need some evidence of positive selection, correlation with disease, and/or a lot of experimental lab work. None of that is even considered in Sternberg’s analysis.
An entirely accurate summary of his argument:
1. We see a pattern in the data.
2. I can’t figure out why its there.
3. Therefore it must be functional.
It is an argument from ignorance, and not even a good one.
I’ve never actually seen Dr. Swamidass’s doctorate, but even though I know it’s not the sign of a good skeptic I accept it on faith.
Dr. Swamidass, you believe God is also constantly tinkering too, right? You just don’t believe that His “tinkering” could possibly leave behind any sign of His presence. Like a burning bush, for example.
Have I got that right?
Welcome to TSZ. We need more theistic evolutionists here!
So the pattern is real, not imagined? Isn’t randomness also a pattern? And we can see this pattern in the data because it deviates from a random pattern. I’m going to assume that answer to that is yes and ask why people are being snarky about Sal’s use of “random.”
Is there some reason patterns cry out for explanation? Isn’t it all just noise?
Mung,
A good skeptic would know that those aren’t the only two options.
Rumraket:
Sal:
Um, I just did. Here it is again:
They aren’t uniformly distributed, but that doesn’t mean they’re non-random.
You learned something, Sal.
Space alien, I think.
Technically, Dr. Swamidass is a (*gasp*) Creationist, and cannot be trusted. By the way, Patrick, atheists have a long history of lying and causing the deaths of untold millions. Not exactly the sort of people I’d want to trust. Just saying.
Dr. Swamidass,
For what it’s worth, I’m not trying to trash the work of Biologos and Francis Collins. Although I’m not a biologist, I am interested in biology and am a part-time grad student of biology at the Foundation for Advanced Education in the Science graduate school at the National Institutes of Health.
It’s very comforting to see Dr. Collins’ prominent position there and when I pass by the book store at the NIH, his book Language of God is always on display. So even though I’m no longer an evolutionist but a Creationist, I’m extremely grateful for your colleague Francis Collins.
I also don’t approve of some of the nasty things some creationists have insinuated of his character. So although I referenced Rick Sternberg’s article, my purpose was not to cast aspersions to biologos, but to discuss the SINE pattern. I posted my thoughts on Dr. Collins here after he visited my undergrad campus and autographed my book:
Some creationists suspect mice and rats share a common ancestor (are from the same created kind), so the question of how the SINE’s have that pattern is not necessarily a claim of special creation. I posted to solicit opinions on the mechanism of the pattern that appears conserved. You suggested a possibl mechanism for non-random insertion preferences:
Thank you for that reference, but I’ve not found any thing that identifies the specific DNA structure targeted for integration by the class of SINES in question (mouse B1/B2/B4, rat ID, primate Alu), at least none that are substantial. I found this however:
Not exactly. I think that God could be tinkering, but it is obvious that none of this tinkering has left a clear signature that He has done so. I suppose He could if He wanted to, but He hasn’t, and it is a silly prior to assume He would.
Well, I do believe God created us through evolution. So if that is what you mean, you are right. Most people, however use “creationist” to mean people that are disputing mainstream science’s account of our origins. I do not.
Scientists do not usually have a problem with people like me. Collins for example fits in this category and is head of the NIH. Tolerance for us is called “strategic ambiguity” by the NSCE and AAAS sometimes.
stcordova,
Sal, I’m pretty sure I acknowledged that some SINEs are functional, and even gave some examples of it. I’m not arguing that all of them are junk.
I would actually argue the opposite. Alu’s in particular seem to have play an important evolutionary role in humans, because they can carry transcriptional elements. So transposon hopping is a really effective way of rapidly (in evolutionary time) modifying genomes. Most transposon hops are safe (because hitting a critical exon is rare, and tinkering with gene expression so much that it kills you is hard), so we can tolerate a high rate without a high deleterious mutational burden. So Alu “functions” as a relatively safe way of rapidly varying gene expression.
Take this with the remarkable fact that (at first approximation) we have essentially the exact same set of proteins (i.e. parts) as chimpanzees, just expressed (i.e. mixed) in different ways. This is one reason many people think Alu has played a very important role in evolution; it functions to “remix” genomes, which (to first approximation) is all you need to generate a human from our common ancestor with apes.
Regardless of how functional or not SINEs are, Sternberg’s argument is really flawed. I don’t have time to pick out the key quotes from that article (sorry, really not trying to be dismissive here), but you can see that several of the patterns are directly explained in that paper, and there are clear pointers to explain the rest. There are, of course, more papers I could point to about the specificity of transposases, that would round out the whole picture. But all this is available on PubMed.
Even if you disagree on specifics, for Sternbergs argument to hold water he has to rule out all known and unknown mechanisms for generating this pattern. Honestly, he has not even tried.
Some people might think that raising a dead man to life would qualify as tinkering and qualify as leaving evidence of said tinkering. 🙂
So God created the universe, life, and evolution. I think that here at TSZ this means you’re a creationist. 🙂
It’s odd though, that you think that what it means to be a creationist depends on modern (“mainstream”) science. imo.
For sure, I do not have a problem with that kind of creationism. I doubt that there is any possible evidence that could show it true or false. So I’m neutral on that question.
Atheist-friendly Creationism. 🙂
Why is that odd?
Shouldn’t it be that a creationist would see science as addressing the question of what was created and how it was created?
I didn’t say non-uniformly distributed necessarily implies they are non-random. You’re attributing arguments to me I didn’t make.
The evidence of non-randomness is that the pattern appears simultaneously in the mouse and rat lineages.
How about “evidence friendly creationism”? I think that’s a better description.
Well yes, I do believe in that, and think God left evidence of His work there. I would hardly characterize this as mere “tinkering.” If Jesus rose from the dead, that changes everything. But what does the Resurrection have to do with evolution and origins? I thought we were talking about creationism and evolution.
Dr. Swamidass,
Thank you very much again for your responses. As I said, there are creationists who think mice and rats share common ancestors, as well as some who think the SINES are junk. There is not uniform agreement.
Ok, since you were insistent, I checked some more.
What I found is there is some preference for certain parts of the DNA, and though I’m not convinced site preference is sufficient to generate the observed pattern in mice and rats, neither can I rule it out.
This Monday I received a request to look into Alu issues partly because of the relatively recent papers suggesting Alu and LINE are functional.
In light of what you say, I can’t in good conscience argue definitively that Sternberg’s observation indicates some sort of common design or intervention. I can only have suspicions, but I don’t think I’d feel comfortable standing by them, hence I felt it was important to raise this discussion again.
I can say however, a good number of researchers (obviously sympathetic to ENCODE) believe a substantial proportion of Alus and LINE-1s have medically significant biological function. Even though they don’t have 100% proof, it’s a good enough working hypothesis which can be further explored. Personally, I think they are functional for the simple intuitive reason that if the human genome is 90% junk, that implies would mean 330,000 base pairs (or 82 megabytes of information) would create something as complex as a human. 82 megabytes seems too small an amount of information to me to create a human…
FWIW, even though I self-identify as an IDist and creationist, I’ve argued it is not appropriate to promote ID as science. The report I’m preparing to circulate in ID circles relates to the more modest question of Alu and LINE function.
Evolutionary biologist Dan Gruar said, “If ENCODE is right, evolution is wrong.” Whether or not that statement is correct, as a purely academic question, it is interesting to me to see if the human genome is mostly functional or not. IDists would naturally side with ENCODE, and I’ve tried to compile evidence to support the ENCODE hypothesis that a large fraction of the human genome is functional. Hence my interest in lncRNAs, SINES, LINES, Alu SINES, etc.
So regarding the SINES in mice and rats, the pattern is non-random, but the papers that explore synthetic transposons (like Sleeping Beauty) integration demonstrate non-random insertion preferences for gene regions.
So even though that doesn’t absolutely account for integration of naturally occurring SINES, it’s enough to suggest this is a possible mechanism, and hence can’t be ruled out as a cause.
I’m very much indebted to you for your constructive criticism. Thank you and God bless you.
How? If Jesus rose from he dead how does that change your theistic evolutionist stance where God does not tinker, or if God does tinker it’s not evident?
We’re talking about whether you are a creationist, because if you are a creationist we cannot assume that you are posting in good faith, even if to do otherwise is to violate the rules of the site. I accept that you are posting in good faith, even if you are a creationist. But there are people here who won’t grant you even that much.
Just trying to warn you. 🙂
Do you have an evolutionary explanation for the resurrection? If not, then you must be one of those utterly despicable “God of the Gaps” theists.
Ok, just took another look.
SINES ( primate Alu, Mouse B1/B2/B4, rat ID) and LINES are Class 1 transposons
So do class I’s use transposase at all????
This is the class listed that uses transposase:
So now I’m really confused! Where is Allan Miller when you need him?
He’s probably all choked up laughing at your incompetence.
Sternberg pointed this out regarding the 2 members of Class 1 transposons: LINES and SINES:
I post the following because
A. LINES and SINES patterns may not be due to transposases
B. “the majority of SINEs integrate at chromosomal breaks by using random DNA breaks to prime reverse transcriptase”
I don’t see the role of transposase in any of this. Someone feel free to help me understand this. Thanks in advance!
I don’t see why the pattern below should be created by transposes at all. If transposases aren’t involved in the first place it’s a moot point. In case they are, then why make the LINES have the opposite density of SINES (or vice versa).
Mung,
I consider him a theistic evolutionist, not a creationist. As a group, theistic evolutionists have a much better record of truthfulness than creationists (and particularly YECs) do.
YECs — particularly those who understand the evidence — have to lie massively to themselves in order to protect their beliefs. They’re denying huge swathes of biology, geology, astronomy, physics, and even linguistics, after all. I suspect this makes it easier for them to lie to others.
Good for you!
In any case, his trustworthiness is irrelevant to my statement about good skeptics:
Mung:
keiths:
Think about it, Mung.
swamidass, to Mung:
And the fact that you don’t really sticks in Mung’s craw. Hence his efforts at shit-stirring and portraying you as a creationist whom we evolutionists are out to get.
Sal,
You claimed that the mutations were non-random. It’s in your OP title…
The Sternberg-Collins Paradox for non-random SINE insertion mutations
…and it’s in your comments:
And:
It isn’t surprising that you got it wrong. You come here to get corrections from people who understand this stuff better than you do, remember?
Sal writes in the OP:
Your description of Dr Sternberg as “one of the most brilliant evolutionary biologists of the present day” induced me to look at Dr Sternberg’s publication record on his website. I’m still wondering why the description? His publications don’t seem all that remarkable at first glance. Have you a link to any recent paper? I can’t see anything more recent than 2005. The Biologic Institute says he “joined Biologic Institute as a principal investigator in 2007” but they don’t list any publications for him as far as I can tell.
Yet Dr Sternberg still works as a “Research Collaborator at the National Museum of Natural History”.